Research
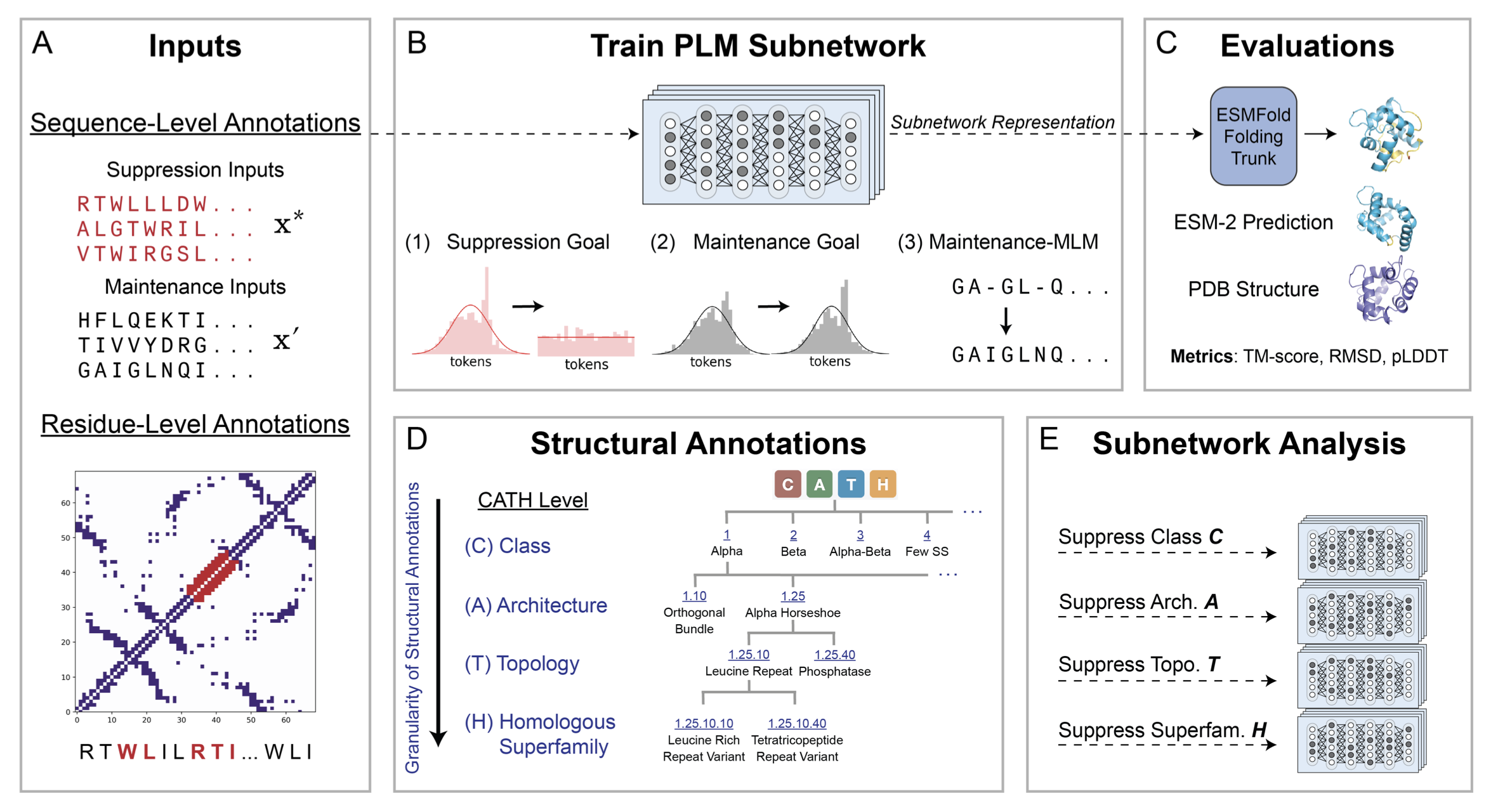
Trainable subnetworks reveal insights into structure knowledge organization in protein language models
Ria Vinod, Ava P. Amini, Lorin Crawford†, Kevin K. Yang†
Paper / Code
Ria Vinod, Ava P. Amini, Lorin Crawford†, Kevin K. Yang†
Paper / Code
bioRxiv, 2025
Subnetwork discovery reveals how secondary structure knowledge is factorized in model weights and how this influences downstream LM-based structure prediction.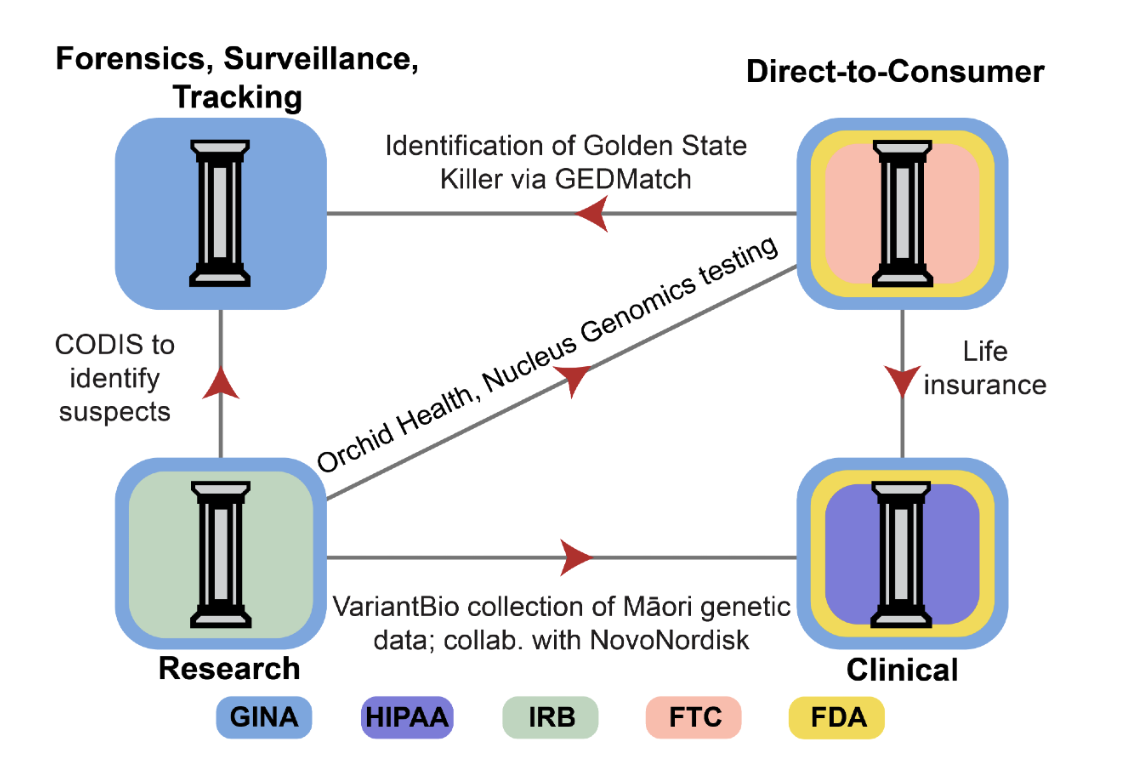
Principles and Policy Recommendations for Comprehensive Genetic Data Governance
Vivek Ramanan*, Ria Vinod*, Cole Williams*, Sohini Ramachandran†, Suresh Venkatasubramanian†
Paper
Vivek Ramanan*, Ria Vinod*, Cole Williams*, Sohini Ramachandran†, Suresh Venkatasubramanian†
Paper
AIES Conference, 2025
A four-pillar risk assessment framework organizes genetic data usage around privacy and ownership principles and guides unified policy reforms spanning legal definitions, GINA coverage, and centralized oversight.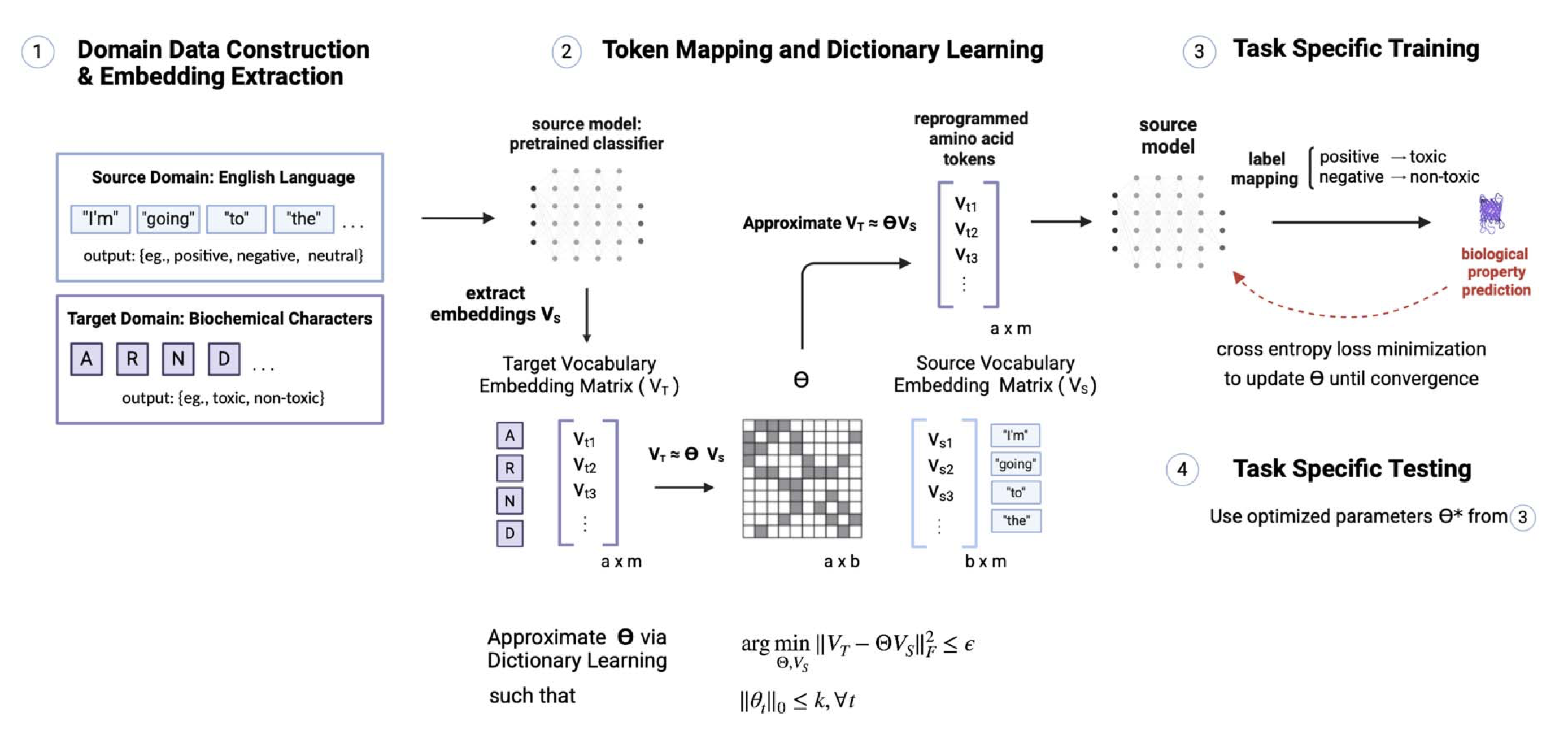
Reprogramming pretrained language models for protein sequence representation learning
Ria Vinod, Pin-Yu Chen, Payel Das†
Paper / Code
Ria Vinod, Pin-Yu Chen, Payel Das†
Paper / Code
RSC Digital Discovery, 2025
A LM-reprogramming framework maps pretrained LM embeddings into protein sequence space, achieving up to 10⁴× data-efficiency gains on molecular learning tasks.Workshops / Other
Logit Subspace Diffusion for Protein Sequence Design
Ria Vinod, Nathan Frey, Andrew Watkins, Jae Hyeyeon Lee
A score-based stochastic differential equation (SDE) framework diffuses over zero-identity component simplexes of protein sequences for antibody design.
Ria Vinod, Nathan Frey, Andrew Watkins, Jae Hyeyeon Lee
A score-based stochastic differential equation (SDE) framework diffuses over zero-identity component simplexes of protein sequences for antibody design.
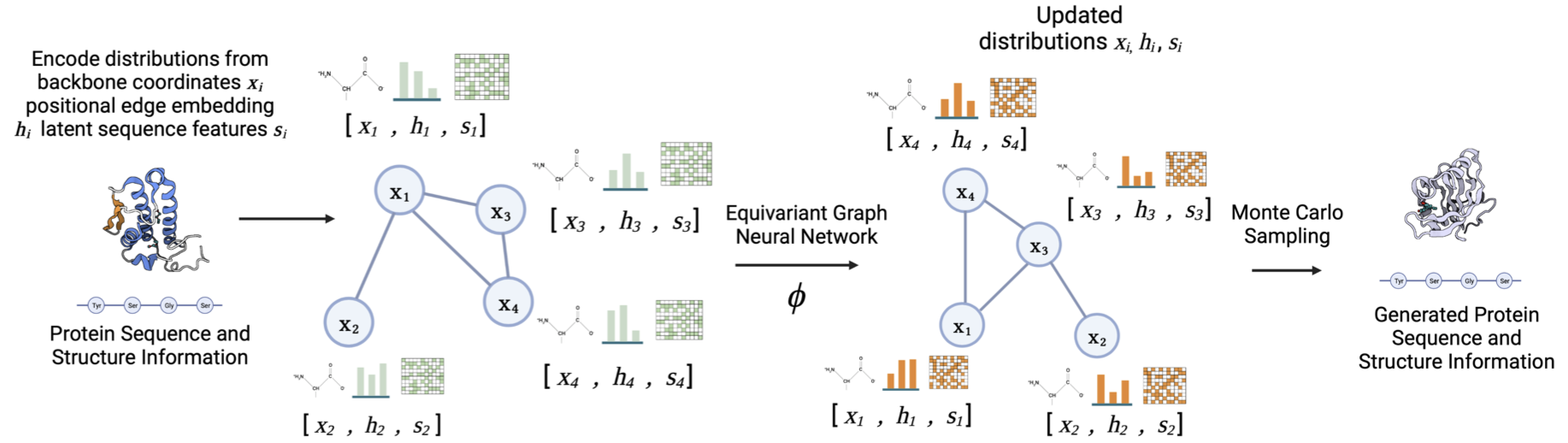
Joint Protein Sequence-Structure Co-Design via Equivariant Diffusion
Ria Vinod, Kevin K. Yang†, Lorin Crawford†
Paper
Ria Vinod, Kevin K. Yang†, Lorin Crawford†
Paper
LMRL @ NeurIPS 2022, Spotlight
A diffusion-based generative modeling framework to probe sequence–structure distribution correlations to improve downstream protein prediction tasks.