
Hi! I’m a 5th-year PhD student at Brown University in the Center for Computational and Molecular Biology. I am fortunate to be advised by Lorin Crawford and to work closely with Kevin Yang and Ava Amini. My PhD research broadly focuses on building methods to gain mechanistic insights into and push the frontiers of biological language models. My academic side quest is to explore how broad bio-sequence collection and subsequent scientific advances interact with society in practice, raising questions of utility, data ownership, and equity.
I completed by B.S. in Electrical Engineering and Computer Science from UC Berkeley. I’ve spent summers working on ML for proteins at Google Deepmind (with Andreea Gaane), Presicient Design (with Jae Hyeon Lee), and IBM Research (with Pin-Yu Chen and Payel Das).
I also spend time co-organizing a Machine Learning x Proteins seminar series and co-run after-school computational science workshops at Providence public schools with some of my wonderful peers.
You can reach me via email.
Research
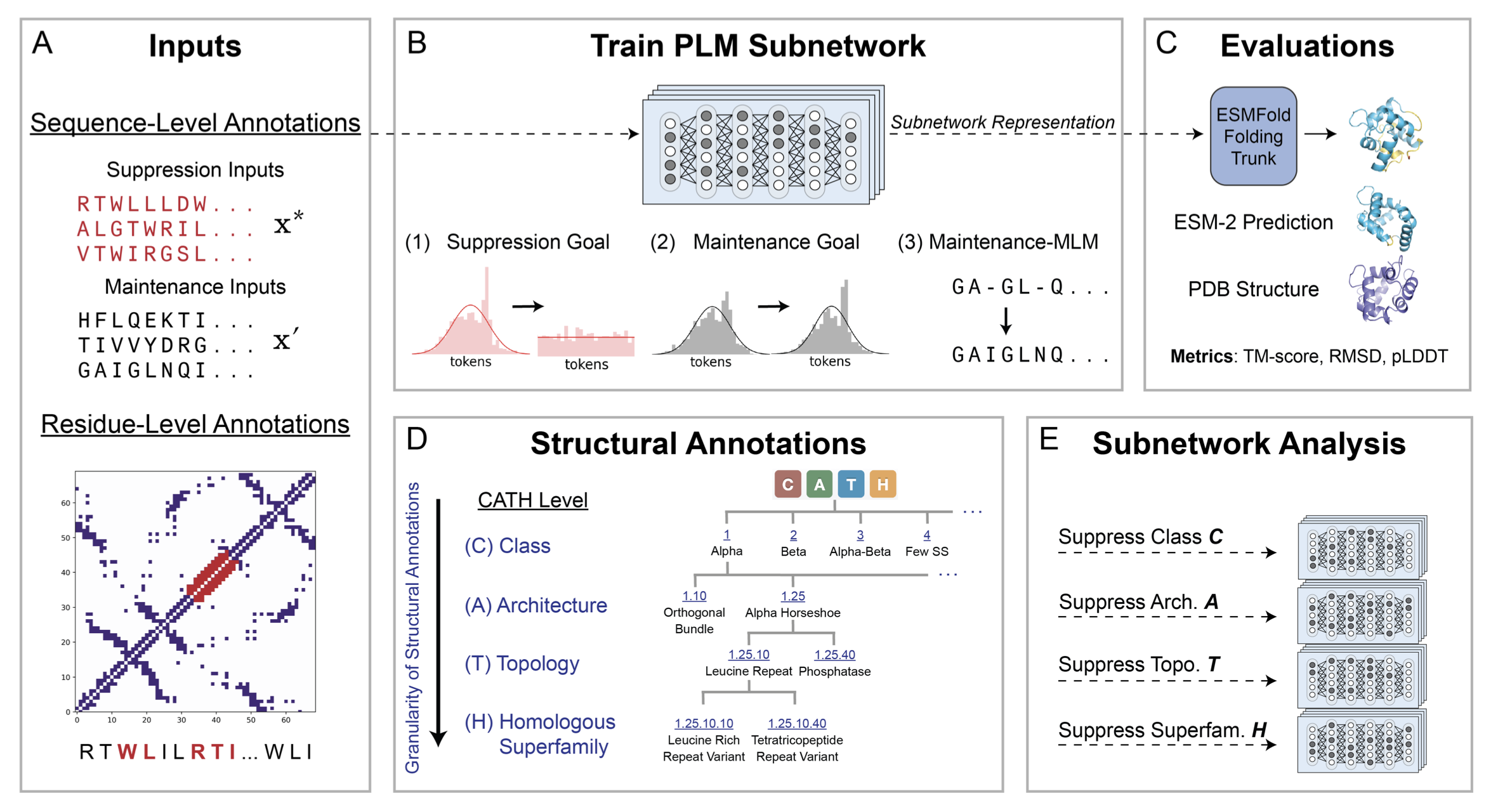
Trainable subnetworks reveal insights into structure knowledge organization in protein language modelsRia Vinod, Ava P. Amini, Lorin Crawford†, Kevin K. Yang†
Paper /
CodebioRxiv, 2025
Subnetwork discovery reveals how secondary structure knowledge is factorized in model weights and how this influences downstream LM-based structure prediction.
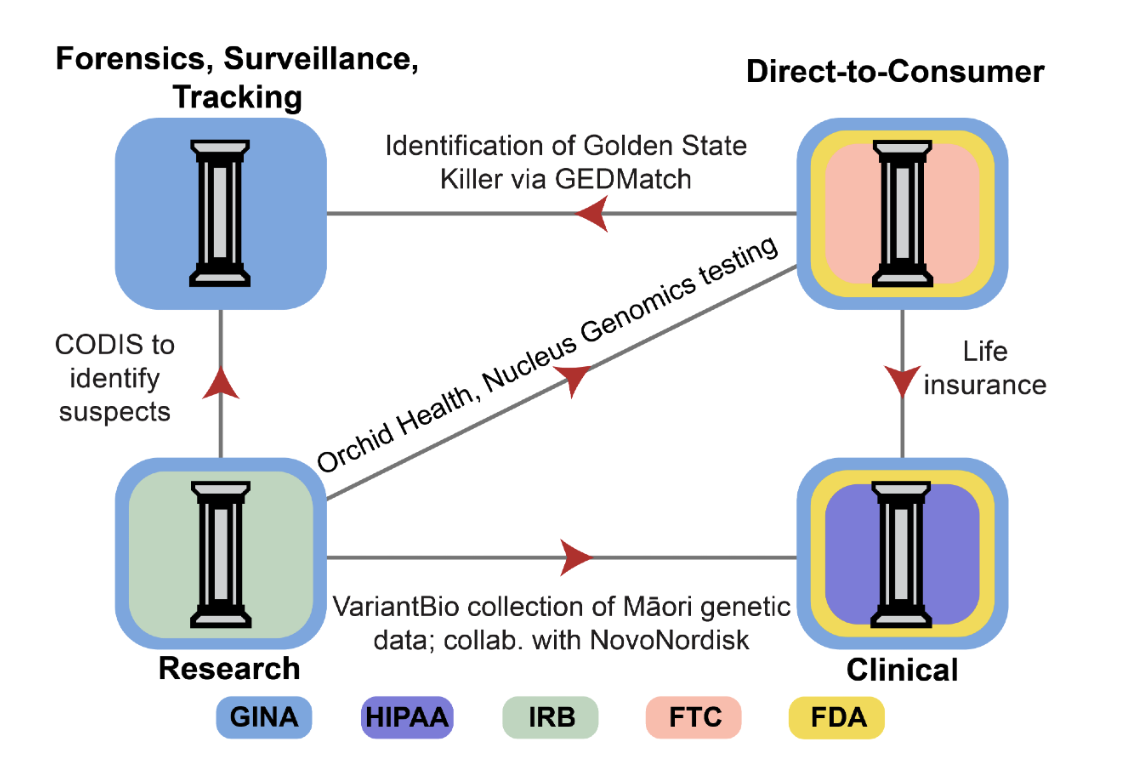
Principles and Policy Recommendations for Comprehensive Genetic Data GovernanceVivek Ramanan*, Ria Vinod*, Cole Williams*, Sohini Ramachandran†, Suresh Venkatasubramanian†
PaperAIES Conference, 2025
A four-pillar risk assessment framework organizes genetic data usage around privacy and ownership principles and guides unified policy reforms spanning legal definitions, GINA coverage, and centralized oversight.
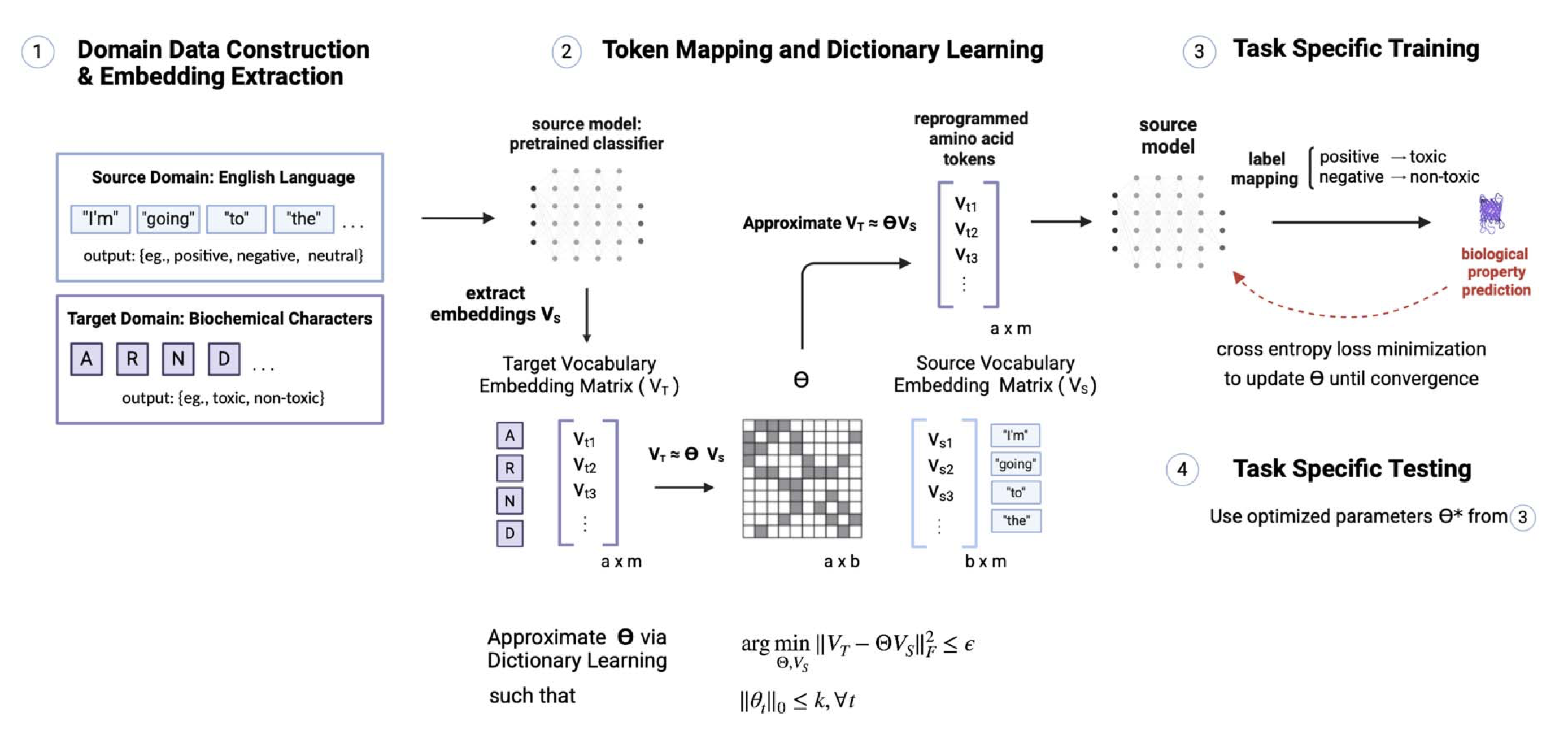
Reprogramming pretrained language models for protein sequence representation learningRia Vinod, Pin-Yu Chen, Payel Das†
Paper /
CodeRSC Digital Discovery, 2025
A LM-reprogramming framework maps pretrained LM embeddings into protein sequence space, achieving up to 10⁴× data-efficiency gains on molecular learning tasks.
Workshops / Other
Logit Subspace Diffusion for Protein Sequence Design
Ria Vinod, Nathan Frey, Andrew Watkins, Jae Hyeyeon Lee
A score-based stochastic differential equation (SDE) framework diffuses over zero-identity component simplexes of protein sequences for antibody design.
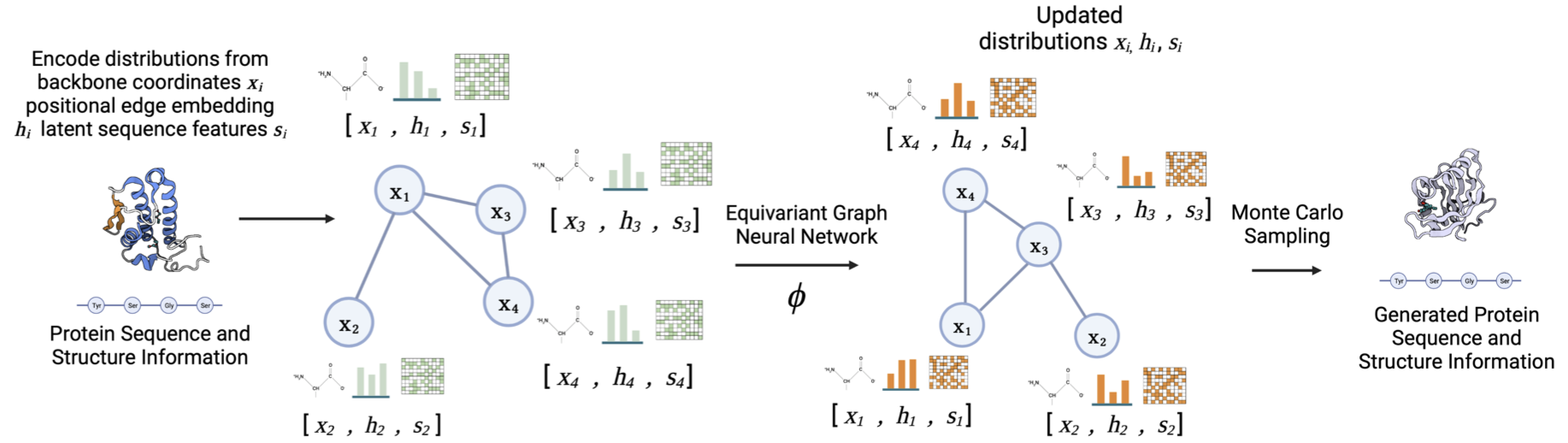
Joint Protein Sequence-Structure Co-Design via Equivariant DiffusionRia Vinod, Kevin K. Yang†, Lorin Crawford†
PaperLMRL @ NeurIPS 2022, Spotlight
A diffusion-based generative modeling framework to probe sequence–structure distribution correlations to improve downstream protein prediction tasks.





